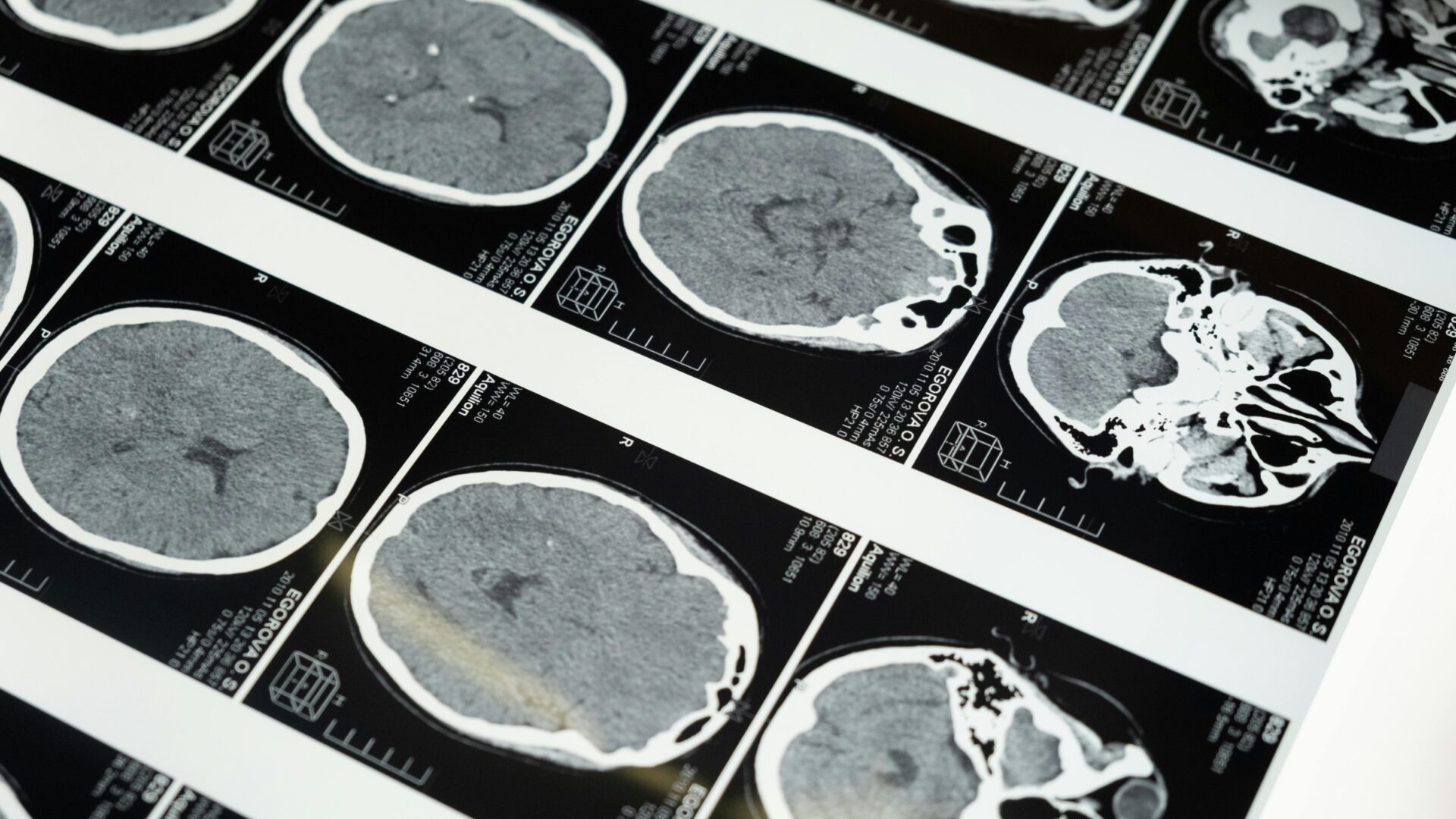
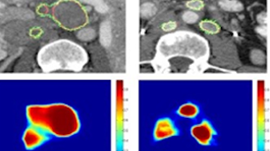
Researchers at UCLA have developed a new AI model that can expertly analyze 3D medical images of diseases in a fraction of the time it would otherwise take a human clinical specialist.
The deep-learning framework, named SLIViT (SLice Integration by Vision Transformer), analyzes images from different imagery modalities, including retinal scans, ultrasound videos, CTs, MRIs, and others, identifying potential disease-risk biomarkers.
Dr. Eran Halperin, a computational medicine expert and professor at UCLA who led the study, said the model is highly accurate across a wide variety of diseases, outperforming many existing, disease-specific foundation models. It uses a novel pre-training and fine-tuning method that relies on large, accessible public data sets. As a result, Halperin believes that the model can be deployed—at relatively low costs—to identify different disease biomarkers, democratizing expert-level medical imaging analysis.
The researchers used NVIDIA T4 GPUs and NVIDIA V100 Tensor Core GPUs, along with NVIDIA CUDA, to conduct their research.
Currently, medical imagery experts are often overwhelmed. Patients often wait weeks to get their X-rays, MRIs, or CT scans evaluated before they can begin treatment.
One of the potential advantages of SLIViT is how it can expertly analyze patient data at scale, and how its expertise can be upgraded. For example, once new medical imaging techniques are developed, the model can be fine-tuned with that new data, which can be pushed out and used in future analyses.
Halperin noted that the model is also easily deployable. Especially in places where medical imagery experts are scarce, in the future the model could potentially make a material difference in patient outcomes.
Before SLIViT, Dr. Halperin said, it was practically infeasible to evaluate large numbers of scans at the level of a human clinical expert. With SLIViT, large-scale, accurate analysis is realistic.
“The model can make a dramatic impact on identifying disease biomarkers, without the need for large amounts of manually annotated images,” Halperin said. “These disease biomarkers can help us understand the disease trajectory of patients. In the future, it may be possible to use these insights to tailor treatment to patients based on the biomarkers found through SLIVIT, and hopefully make a dramatic improvement in patients’ lives.”
According to Dr. Oren Avram, lead author of a paper the UCLA researchers published in Nature Biomedical Engineering, the study revealed two surprising—yet related—results.
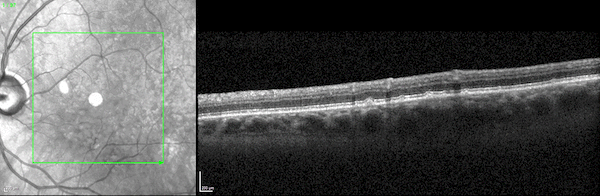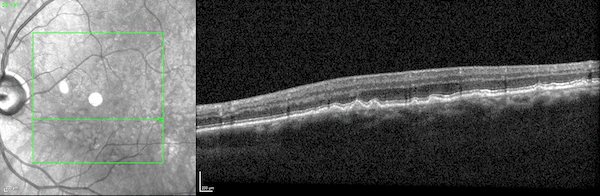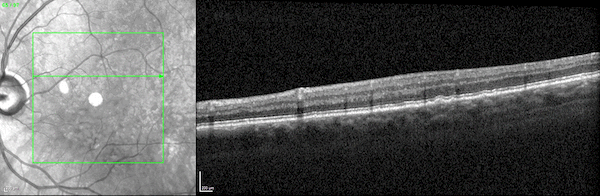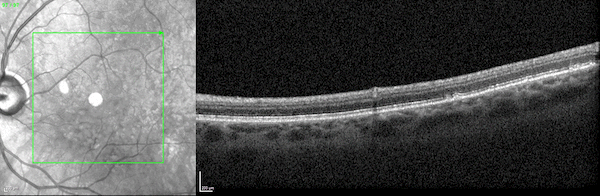
First, while the model was largely pre-trained on datasets of 2D scans, it accurately identifies disease biomarkers in 3D scans of human organs. Typically, a model designed to analyze 3D images is trained on 3D datasets. But 3D medical data is far more expensive to acquire and thus far less abundant and accessible than 2D medical data.
The UCLA researchers found that by pre-training their model on 2D scans—which are far more accessible—and fine-tuning it on a relatively small amount of 3D scans, the model outperformed specialized models trained only on 3D scans.
The second unanticipated outcome was how good the model was at transfer learning. It learned to identify different disease biomarkers by fine-tuning on datasets consisting of imagery from very different modalities and organs.
“We trained the model on 2D retinal scans—so images of your eye—but then fine-tuned the model on an MRI of a liver, which seemingly have no connection, because they’re two totally different organs and imaging technologies,” Avram said. “But we learned that between the retina and the liver, and between an OCT and MRI, some basic features are shared, and these can be used to help the model with downstream learnings even though the imagery domains are totally different.”
Read additional news from UCLA about SLIViT.
Check out the SLIViT paper, Accurate prediction of disease-risk factors from volumetric medical scans by a deep vision model pre-trained with 2D scans.
Access the model on GitHub.